Study of Xylose as Product Inhibitor in Xylanase from Aspergillus niger, Basillus subtilis, and Tricodherma reesei: Insilico and Experimental Review Approach
Abstract
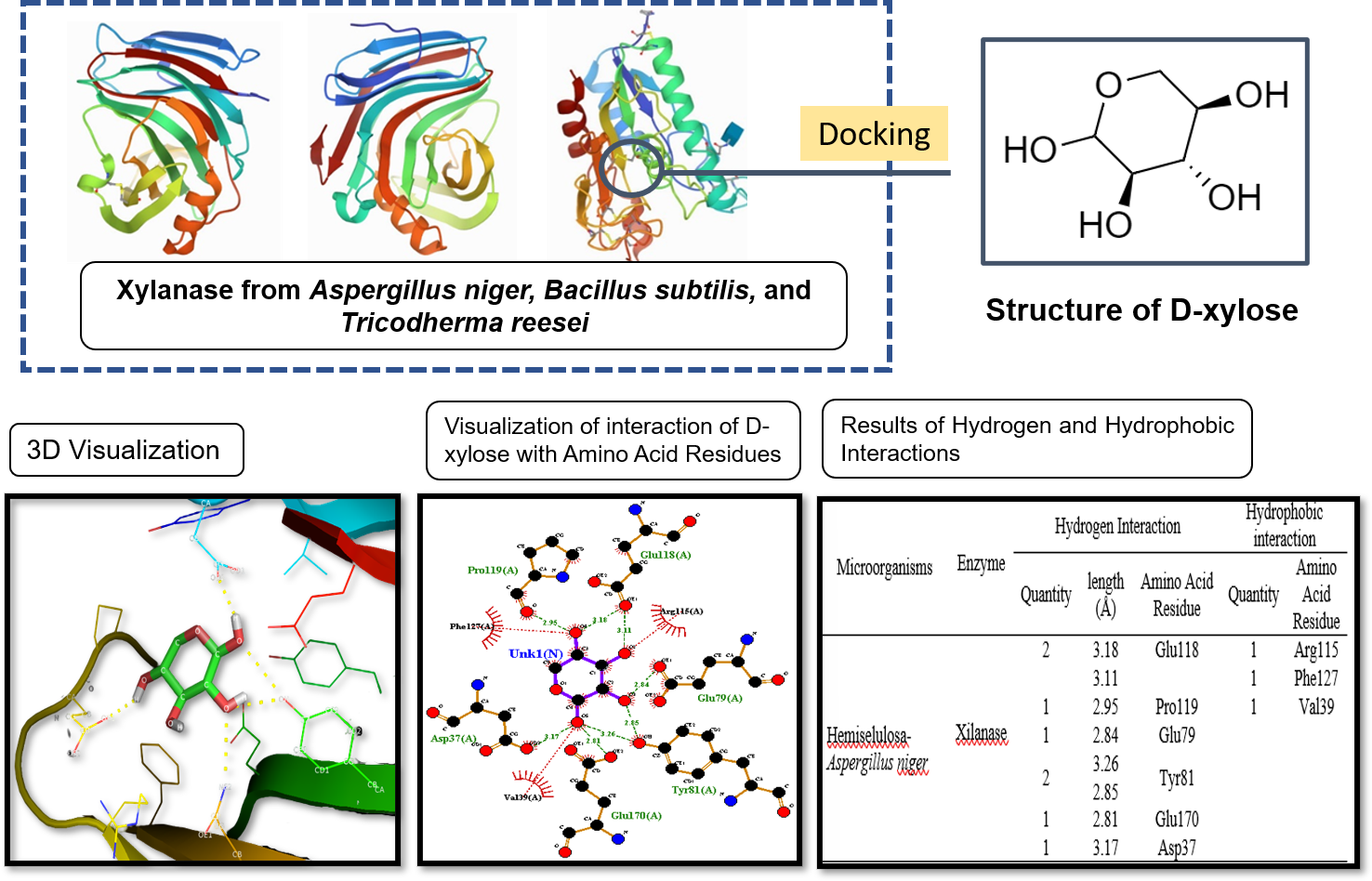
Bioinformatics is useful for solving molecular biology problems computationally. This computational chemistry has the advantage of being safe, free from chemical waste, secure, cost-effective, and can shorten research time. The issues that arise can be in the form of basic things such as solving enzyme mechanisms, protein metabolism, or identifying microbes. Degradation of the xylanase enzyme using some microorganisms. They are Aspergillus niger, Bacillus subtilis, and Thricodherma reesei on lignocellulose bonds. Lignocellulose consists of lignin, cellulose, and hemicellulose. Cellulose and hemicellulose can have used to produce new products such as bio-based products. To predict the optimum conditions for this enzymatic reaction has used bioinformatics applications has used through substrate enzymes obtained from protein data banks. The purpose of this study was to determine the optimum conditions for obtaining xylanase enzymes from the microorganisms Aspergillus niger, Bacillus subtilis, and Thricodherma reesei by bioinformatics (in silico). This research was conducted in bioinformatics using a database from the RCSB Protein Data Bank and PubChem. The programs used to see the interaction between substrate enzymes in this study are PyMol, PyRx, and LigPlot. The best conditions based on the results of bioinformatics simulations will form the basis for producing xylanases on a laboratory scale. In this study, the results of interaction data between Bacillus subtilis and D-xylose, which have a binding affinity value of -5.2 kcal/mol. Aspergillus niger with D-xylose, which has a binding affinity value of -5.1 kcal/mol, Tricodherma reesei with D-xylose, which has binding affinity value -4.3 kcal/mol.
References
[1] Budiman, and Setyawan, S. Pengaruh Konsentrasi Substrat, Lama Inkubasi dan pH Dalam Proses Isolasi Enzim Xylanase Dengan Menggunakan Media Jerami Padi, 2009, 1–11.
[2] Anindyawati, T. Jurnal Selulosa, 2010, 45 (02).
[3] Wang, J., Chen, X., Chio, C., Yang, C., Su, E., Jin, Y., Cao, F and Qin, W, Bioresour. Technol, 2019, 274, 459–467.
[4] Devi, D., Astutik, D., Cahyanto, M. N and Djaafar, T. F, JRPB, 2019, 7 (2), 273–282.
[5] Haliza, W., Sukasih, E and Agustinisari, I. JPASCA, 2007, 4 (1), 9–17.
[6] Ottenheim, C., Verdejo, C., Zimmermann, W and Wu, J. C. J. Biosci. Bioeng, 2014, 118 (6), 696–701.
[7] Sutrisno, S., Roosdiana, A., Prasetyawan, S and Sari, I. P, JKPK (Jurnal Kimia dan Pendidikan Kimia), 2017, 2 (2), 97–102.
[8] Iyyappan, J., Bharathiraja, B., Baskar, G and Kamalanaban, E, Bioresour. Technol, 2019, 281, 18–25.
[9] Jayus, J., Nafi’, A and Hanifa, A. S, J-AGT (Jurnal Agroteknologi), 2019, 13 (01), 34-41.
[10] Parikesit, A. A, The Role of Bioinformatics as Auxilliary Tools for Molecular Biology,” Oct. 2009.
[11] Can, T, Methods. Mol. Biol, 2014, 1107, 51–71.
[12] Nugraha, W., Suwartawan, W., Prayoga, A., Laksmiani, L., Putra, P and Ani, S, Jurnal Farmasi Udayana, 2018, 1–6.
[13] Azam, S. S and Abbasi, S. W., Theor. Biol. Medical Modelling, 2013, 10 (1), 1-16.
[14] Schmidt, T and Schlegel, H. G., J. Bacteriol, 1994, 176 (22), 7045–7054.
[15] Dashek, W. V, Methods in Plant Biochemistry and Molecular Biology, (1st ed.)., 1997, CRC Press.
[16] Haltrich, D., Nidetzky, B., Kulbe, K. D., Steiner, W and Župančič, S, Bioresour. Technol, 1996, 58 (2), 137–161.
[17] Trott, O and Olson, A. J, J. Comput. Chem, 2010, 31 (2), 455–461.
[18] Maryanty, Y and Sumitro, S. B., Interaction of Enzyme-Substrate from Indigenous Cellulolytic Bacteria by Bioinformatics. IOP Conf. Ser: Mater. Sci. Eng., 2020, 854 (1), 012068.
[19] Plácido, J and Capareda, S, Bioresources and Bioprocessing, 2015, 2 (1), 1-12.
[20] Chen, M., Zeng, G., Tan, Z., Jiang, M., Li, H., Liu, L., Zhu, Y., Yu, Z., Wei, Z., Liu, Y and Xie, G. PLOS ONE 2011, 6 (9), e25647.
[21] Tim INBIO Indonesia. Cara Mudah Melakukan Docking Dengan PyRx (Autodock VINA), Global Sciense.
[22] Selvam, K., Senbagam, D., Selvankumar, T., Sudhakar, C., Kamala-Kannan, S., Senthilkumar, B and Govarthanan, M. J. Mol. Struct, 2017, 1150, 61–67.
[23] Arwansyah, A., Ambarsari, L., Sumaryada, T. I. Curr. Biochem. 2014, 1 (1), 11–19.
[24] Rasyidi, A. S., Studi Potensi Beberapa Senyawa yang Memiliki Aktivitas Antiobesitas dengan Metode Penambatan Molekul, 2018.
[25] Rawat, R., Kumar, S., Chadha, B. S., Kumar, D and Oberoi, H. S. Antonie van Leeuwenhoek, 2015, 107 (1), 103–117.
[26] Nauli, T. Jurnal Kimia Terapan Indonesia, 2014, 16 (2), 94–100.
[27] Aziz, F. K., Nukitasari, C., Oktavianingrum, F. A., Aryati, L. W., Santoso, B. Jurnal Kimia VALENSI, 2016, 2 (2) 120–124.
[28] Pratama, R. Penambatan Molekuler Senyawa Aktif Temulawak (Curcuma xanthoriza) Dengan Enzim Cox-2 SebagaiKandidat Obat Anti Kanker Payudara. Thesis, Bogor Agricultural University (IPB), 2015.Refbacks
- There are currently no refbacks.

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.