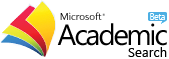The Discovery of Tyrosinase Enzyme Inhibitors Activity from Polyphenolic Compounds in Red Grape Seeds through In Silico Study
Abstract
References
[1] Hoogduijn, M.J., Cemeli, E., Ross, K., Anderson, D., Thody, A.J., and Wood, J.M., Exp. Cell Res., 2004, 294(1), 60-67.
[2] Chang, T.S., Materials., 2012, 5(9), 1661-1685.
[3] Ghanem, G., and Fabrice, J., Mol. Oncol. 2011, 5(2), 150-155.
[4] Olivares, C., and Solano, F., Pigment Cell and Melanoma Research., 2009, 22(6), 750-760.
[5] Briganti, S., Camera, E., and Picardo, M., Pigment Cell Res., 2003, 16(2), 101-110.
[6] Vashi, N.A., and Kundu, R.V., Br. J. Dermatol., 2013, 169, 41-56.
[7] Jiang, J., Akinseye, O., Tovar-Garza, A., and Pandya, A.G., IJWD., 2018, 4(1), 38-42.
[8] Darji, K., Varade, R., West, D., Armbrecht, E.S., Guo, M.A., JCAD., 2017, 10(5), 18-23.
[9] Panich, U., Current Management of Malignant Melanoma., 2011, 227.
[10] Manach, C., Williamson, G., Morand, C., Scalbert, A., and Rémésy, C., Am. J. Clin. Nutr., 2005, 81(1), 230S-242S.
[11] Zhu, F., Du, B., and Li, J., Int. J. Wine. Res. 2015, 7, 63-67.
[12] Goldfeder, M., Kanteev, M., Isaschar-Ovdat, S., Adir, N., and Fishman, A., Nat. Commun., 2014, 5(1), 1-5.
[13] Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R, et al. Gaussian09 Revision D.01, Gaussian Inc. Wallingford CT. Gaussian 09 Revision C.01. 2010.
[14] Goodsell, D.S., Morris, G.M., and Olson, A.J., J. Mol. Recognit., 1996, 9(1), 1-5.
[15] Morris, G.M., Goodsell, D.S., Halliday, R.S., Huey, R., Hart, W.E, Belew, R.K., et al. J. Comput. Chem., 1998, 19(14), 1639-1662.
[16] Humphrey, W., Dalke, A., and Schulten, K., J. Mol. Graph. 1996, 14(1), 33-38.
[17] Pronk, S., Páll, S., Schulz, R., Larsson, P., Bjelkmar, P., Apostolov, R., et al. Bioinformatics., 2013, 29(7), 845-854.
[18] Abraham, M.J., Murtola, T., Schulz, R., Páll, S., Smith, J.C., Hess, B., and Lindahl, E., SoftwareX. 2015, 1, 19-25.
[19] Páll, S., Abraham, M.J., Kutzner, C., Hess, B., and Lindahl, E., Tackling exascale software challenges in molecular dynamics simulations with GROMACS In: Solving Software Challenges for Exascale. EASC 2014. Lecture Notes in Computer Science. Solving Softw Challenges Exascale. 2015, 3-27.
[20] Aliev, A.E., Kulke, M., Khaneja, H.S., Chudasama, V., Sheppard, T.D., and Lanigan, R.M., Proteins, 2014, 82(2), 195-215.
[21] Da Silva, A.W.S., and Vranken, W.F., BMC Res Notes. 2012, 5(1), 1-8.
[22] Darden, T., York, D., and Pedersen, L., J. Chem. Phys. 1993, 98(12), 10089-10092.
[23] Essman, U., Perera, L., Berkowitz, M.L., Darden, T., Lee, H., and Pedersen, L.G., J. Chem. Phys. 1995, 103, 8577.
[24] Bhhatarai, B., Wilson, D.M., Parks, A.K., Carney, E.W., and Spencer, P.J., Chem Res Toxicol. 2016, 29(5), 810-822.
Refbacks
- There are currently no refbacks.

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.