Profiling of The Lemongrass Oil Aroma and Their Structure-Odor Relationship: In Silico Study
Abstract
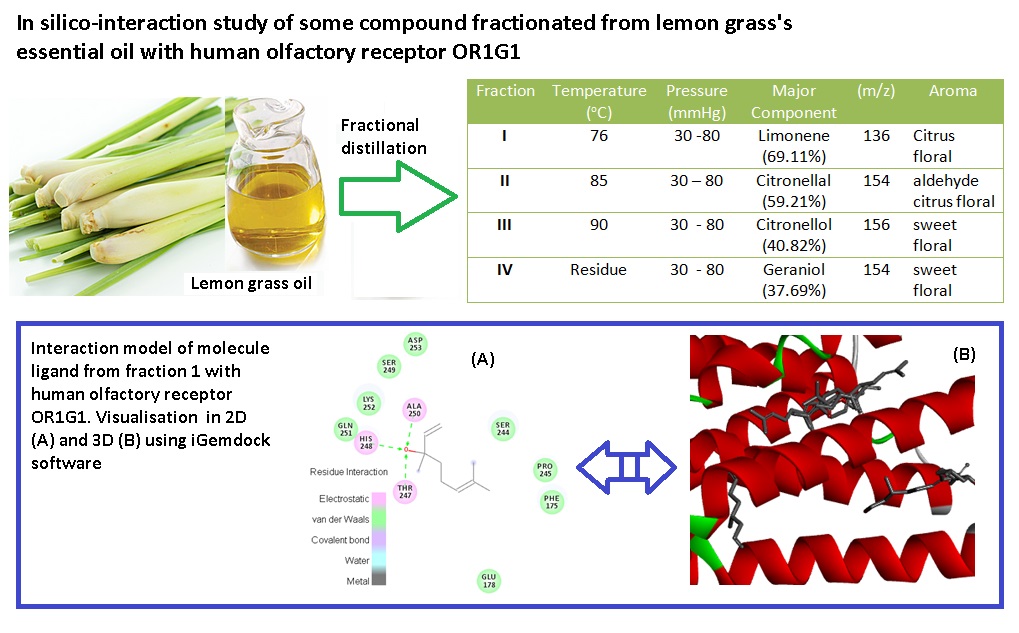
Structure-odor relationship (SOR) has previously studied by semantic numerically in different Fragrance. We hypothesise that in silico method such as molecular dynamics, together with docking of the interaction between human olfactory receptor (OR1G1) and ligands, can offer extremely valuable tools of modelling SOR. Hence, the present study was carried out to express the SOR of citronellal oil fraction compare with reference smelling of floral, musk, green, wood, and fruit by employing docking and multiple discriminant analysis (MDA). Our study reveals that the number dissociation constant (Kd), bond distance, HOMO-LUMO (AE), dipole moment, kind of amino acids, Log P, surface area and hydropathy as the variable SOR from in silico anaysis. Our result has shown ligands and OR1G1 interacted with Van Der Waals and electrostatic model. MDA analysis shown molecule reference floral and fraction of lemongrass oil have similar correlation based on variable SOR with linier regression of all variable SOR to Kd value for every reference odor is R2 = 1.
References
[1] K. Sucker, R. Both, M. Bischo, R. Guski, U. Krämer, and G. Winneke, Int Arch Occup Environ Health., 2008, 81(6), 683–694.
[2] K. J. Rossiter, E. Fruit, and H. Wood, Chem. Rev., 1996, 96 (8), 3201–3240.
[3] L. Turin, Structure-odor relations : a modern perspective in Handbook of olfaction and gustation, 1991, Wiley-Blackwell, 1–35.
[4] K. Harini and R. Sowdhamini, PLoS One, 2015, 1–30.
[5] L. Buck and R. Axel, Cell, 1991, 65, 175–187.
[6] P. Mombaerts, Science, 1999, 286(5440), 707-711.
[7] B. Malnic, J. Hirono, T. Sato, L. B. Buck, and H. Hughes, Cell, 2000, 96, 713–723.
[8] K. Audouze, A. Tromelin, A. M. L. Bon, C. Belloir, R. K.Petersen, K. Kristiansen, S. Brunak, O. Tabourea, PLoS One, 2014, 9(4), 1-12.
[9] M. Zarzo and D. T. Stanton, Chem. Senses., 2006, 31(8), 713–724.
[10] K. Mori, Y. K. Takahashi, K. E. I. M. Igarashi, and M. Yamaguchi, Physiol Rev, 2006, 86(2), 409–433.
[11] G. Launay, F. Wade, and E. Pajot-augy, Protein Eng Des Sel, 2012, 25(8), 377–386.
[12] F. Chen and O. Martín-belloso, HANDBOOK OF FRUIT AND VEGETABLE FLAVORS Edited by Y.H. Fuyi, 2010, John Wiley & Sons, Inc.
[13] Moncrieff, R. W., The chemical senses,3d ed. Cleveland, 1967, CRC Press.
[14] Kyte, Jack dan Doolittle, R. F., J. Mol. Biol., 1982, 157, 105-132.
[15] Morris, G. M., R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D S. Goodsell, A. J. Olson, J. Comput. Chem., 30(16), 2785-2791.
[16] Brozell, S. R., S. Mukherjee, T. E. Balius, D. R. Roe, D. A. Case, R. C. Rizzo, J Comput Aided Mol Des, 2012, 26, 749-773.
[17] Hsu, K.-C., Y.-F. Chen, S.-R. Lin and J.-M. Yang, BMC Bioinformatics, 2011, 12, 1-11.
Refbacks
- There are currently no refbacks.

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.