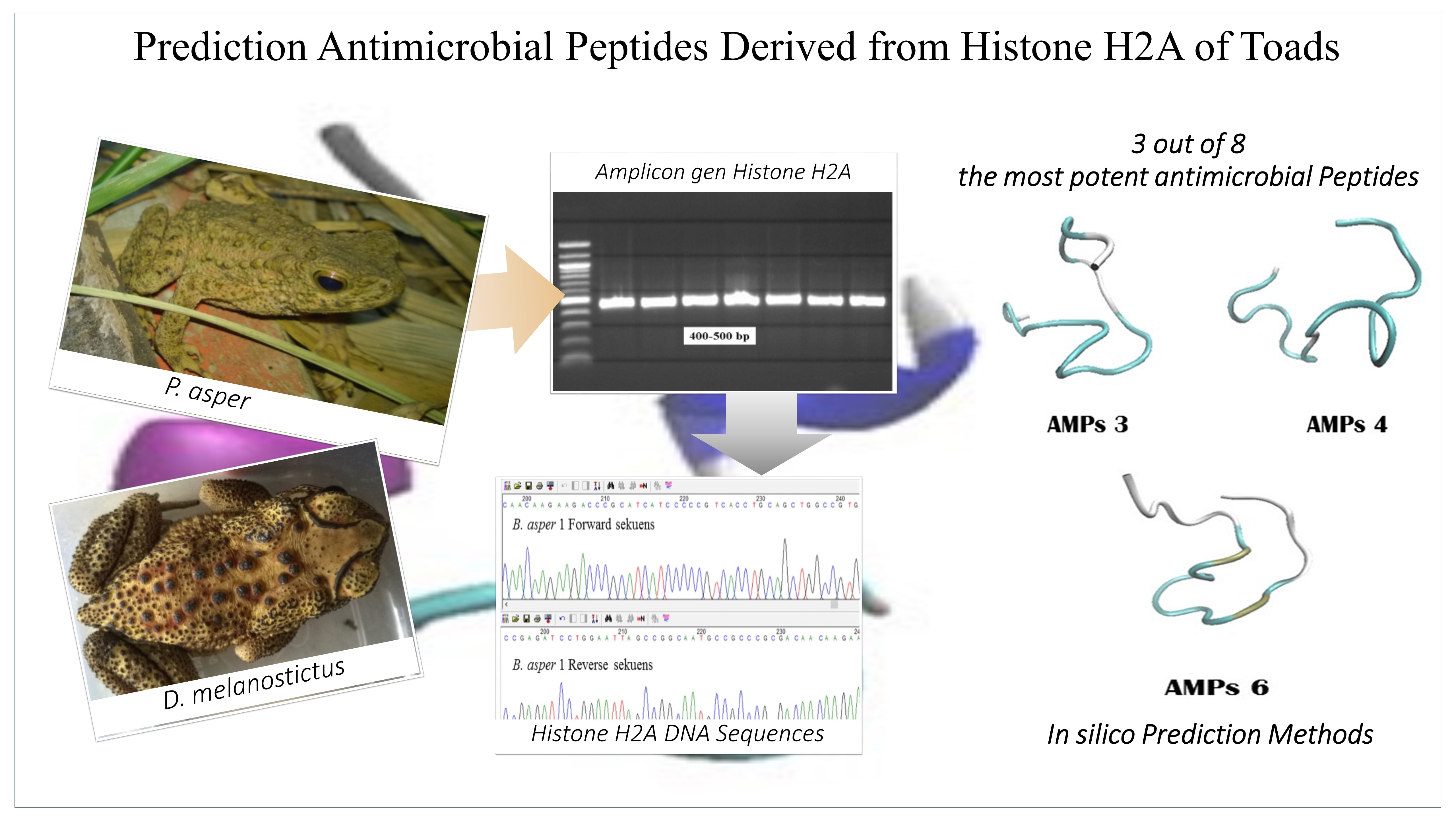
Analysis and Prediction of some Histone-derived Antimicrobial Peptides from Toads Duttaphrynus melanostictus and Phyrinoidis asper
Abstract
References
[1] Song JH., Antimicrobial Resistance Control in Asia, Monitoring, in Carlet J, AMR Control 2015 overcoming Global Antimicrobial Resistance, Surveillance and National Plans, 2015, TIM PROBART Publisher, United Kingdom.
[2] Singh N., Rai V., Bioinformation, 2012, 8(21), 1021-1025.
[3] Sang Y., Blecha F., Anim Health Res Rev, 2008, 9(2), 227-235.
[4] Bulet P., Stocklin R., Menin L., Immunol Rev, 2004, 198, 169-184.
[5] Xu X., Lai R., Chem Rev, 2015, 115(4), 1760-1846.
[6] Zardini HZ., Ebrahimi L., Ejtehad MM., HashemiA., Azam AG., Atef A., Soleimanizadeh M., Turki J Biochem, 2013, 38(4), 416-424.
[7] Garg AD., Kanitkar DV., Hippargi RV., Gandhare AN., Nat Proceed, 2007.
[8] Utami E., Kusumorini A., Anggadiredja K., Barlian A., Proceed 3rd ICMNS, 2010.
[9] Kawasaki H., Iwamuro S., Infect Dis-Drug Targ, 2008, 8, 195-205.
[10] Almaali AHM., IJRSR, 2014, 5(3), 649-655.
[11] Shatyan N., Philip R., Chaithanya ER., Kumar PRA., Sanjeevan VN., Singh ISB., ISRN Mol Biol, 2012, 1-6.
[12] Chaithanya ER., Philip R., Sathyan N., Kumar PRA., ISRN Mol Biol, 2013, 1-7.
[13] Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T., BMC Bioinformatics, 2012, 13(134), 1-11.
[14] Kim HS., Park CB., Kim MS., Kim SC., Biochem Biophys Res Comm, 1996, 229(1814), 381–387.
[15] Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., Mol Biol Evol, 2011, 28, 2731-2739.
[16] Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden TL., BMC Bioinformatics, 2009, 10(421), 1-9.
[17] Yoo WG., Lee MR., Yun T., Kwon., Kim DW., Bioinformation, 2015, 11(1), 017-020.
[18] Wagh FH., Gopi L., Barai RS., Ramteke P., Nizami B., Idicula-Thomas S., Nucleic Acids Res, 2013, 42(1), D1154- D1158.
[19] Rice P., Longden I., Bleasby A., Trends Genet, 2000, 16(6), 276-277.
[20] Fernndez-Escamilla AM., Rousseau F., Schymkowitz J., Serrano L., Nat Biotech. 2004, 22(10), 1302-1306.
[21] Conchilo-Solé O., de-Groot NS., Avilés FX., Vendrell J., Daura X., Ventura S., BMC Bioinformatics, 2007, 8(65), 1-16.
[22] Gasteigr E., Hoogland C., Gattiker A., Duvaud S., Wilkins MR., Appel RD., Bairoch A., Protein Identification and Analysis Tools on the ExPASy Server, in The Proteomics Protocols Handbook, Humana Press. New Jersey, 2005, 571-607.
[23] Kaur H., Garg A., Raghava GPS., Protein Peptides Let, 2007, 14(7), 626-630.
[24] Humphrey W., Dalke A., Schulten K., J Mol Graphics, 1996, 14(1): 33-38.
[25] Nei M., Kumar S., Molecular Evolution and Phylogenetic, 2000, 147-164.
[26] Shai Y., Biopolymers Pept Sci, 2002, 66, 236–248.Refbacks
- There are currently no refbacks.