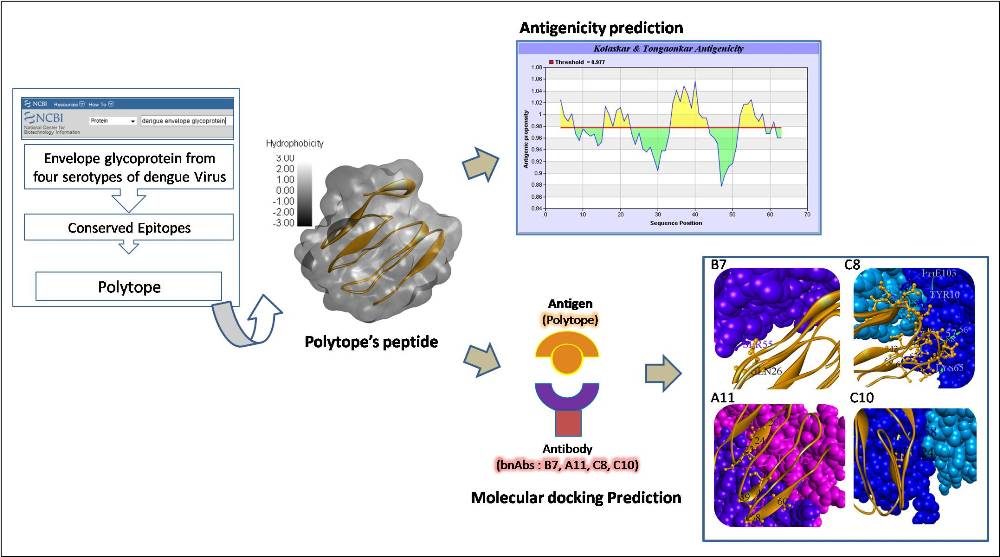
Polytope Prediction for Dengue Vaccine Candidate Based on Conserved Envelope Glycoprotein of Four Serotypes of Dengue Virus and Its Antigenicity
Abstract
References
[1] Amâncio, F. F.; Ferraz, M. L.; Cristina, M.; Almeida, D. M.; Eduardo, J.; Pessanha, M.; Iani, F. C. M.; Fraga, G. L.; Roberto, J.; Carneiro, M. J. Infect. Public Health 2014, 7 (6), 547–552.
[2] Sofia, H.; Monteiro, M. T. T.; Torres, D. F. M. Oper. Res. Heal. Care 2015, 7, 122–131.
[3] Lardo, S.; Utami, Y.; Yohan, B.; Tarigan, S. M. M. U.; Santoso, W. D.; Nainggolan, L.; Sasmono, R. T. Asian Pac. J. Trop. Med. 2016, 9 (2), 134–140.
[4] Myat, M.; Tun, N.; Kyaw, A.; Makki, N.; Muthugala, R.; Nabeshima, T.; Zin, K.; Morita, K. Infect. Genet. Evol. 2016, 43, 31–37.
[5] Murrell, S.; Wu, S.; Butler, M. Biotechnol. Adv. 2011, 29 (2), 239–247.
[6] Ba, A. T. Infect. Ecol. Epidemiol. 2013, 1, 1–22.
[7] Johnson, J.; Hunt, A. N. N. R.; Bolin, A.; Roehrig, J. T.; Chu, M. A. Y. C. Virology 1990, 675, 668–675.
[8] Mart, J.; Hermida, L.; Alvarez, M.; Vald, I.; Chinea, G.; Rosario, D.; Guill, G.; Guzm, G. Virus Res. 2006, 121, 65–73.
[9] Kelly, E. P.; Puri, B.; Sun, W.; Falgout, B. Vaccine 2010, 28 (17), 3030–3037.
[10] Rantam, F. A.; Soegijanto, S.; Susilowati, H.; Sudiana, K.; Hendrianto, E. Trials Vaccinol. 2013, 1–5.
[11] Hunsawong, T.; Sunintaboon, P.; Warit, S.; Thaisomboonsuk, B.; Jarman, R. G.; Yoon, I.; Ubol, S.; Fernandez, S. Vaccine 2015, 33 (14), 1702–1710.
[12] Sánchez-burgos, G.; Ramos-casta, J.; Cedillo-rivera, R.; Dumonteil, E. Virus Res. 2010, 153, 113–120.
[13] Cedillo-barr, L.; Garc, J.; Guti, B. Microbes Infect. 2014, 16.
[14] Venkatachalam, R.; Subramaniyan, V. Asian Pacific J. Trop. Dis. 2015, 5 (Suppl 1), S47–S50.
[15] Paul, William E., M. Fundamental Immunology. Fifth Edition; Lippincott Williams & Wilkins: Maryland, 2003.
[16] Suhrbier, A. Expert Rev. Vaccines 2002, 207–213.
[17] Han, K.; Zhao, D.; Liu, Y.; Huang, X.; Yang, J.; Liu, Q.; An, F.; Li, Y. Res. Vet. Sci. 2016, 104, 174–180.
[18] Fitriyah. Insilico Analisys of Multi-Epitope Vaccine Candidat Design Againts Mannose Receptor to Inhibit Dengue Infection, University of Brawijaya, Malang, 2012.
[19] Zhang, Y. 2008, 8, 1–8.
[20] Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. Nat. Publ. Gr. 2015, 12 (1), 7–8.
[21] Kolaskar, A.; Tongaonkar, P. FEBS Lett 1990, 276 (1-2), 172–174.
[22] Kim, Y.; Ponomarenko, J.; Zhu, Z.; Tamang, D.; Wang, P.; Greenbaum, J.; Lundegaard, C.; Sette, A.; Lund, O.; Bourne; Nielsen, M.; Peters, B. Nucleic Acids ResNucleic Acids Res 2012, 40.
[23] Saha, S.; Raghava, G. P. . Proteins Struct, Funct Bioinf. 2006, 65.
[24] Darnell, S. J.; Page, D.; Mitchell, J. C. Proteins 2007, 68 (4), 813–823.
[25] Zhu, X.; Mitchell, J. C. Proteins 2011, 79 (9), 2671–2683.
[26] Duhovny, D.; Nussinov, R.; Wolfson, H. Algorithms Bioinforma. 2002, 185–200.
[27] Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H. J. Nucleic Acids Res. 2005, 33 (SUPPL. 2), 363–367.
[28] Lesk, A. M. Introduction to Bioinformatics; Oxford University Press: UK, 2002.
[29] Rouvinski, A.; Guardado-Calvo, P.; Barba-Spaeth, G.; Duquerroy, S.; Vaney, M.; Kikuti, C. M.; Sanchez, M. E. N.; Dejnirattisai, W.; Wongwiwat, W.; Haouz, A.; Girard-Blanc, C.; Petres, S.; E.Shepard, W.; Despres, P.; Arenzana-Seisdedos, F.; Dussart, P.; Mongkolsapaya, J.; R.Screato, G.; Rey, F. A. Nature 2015, 1–16.
Refbacks
- There are currently no refbacks.